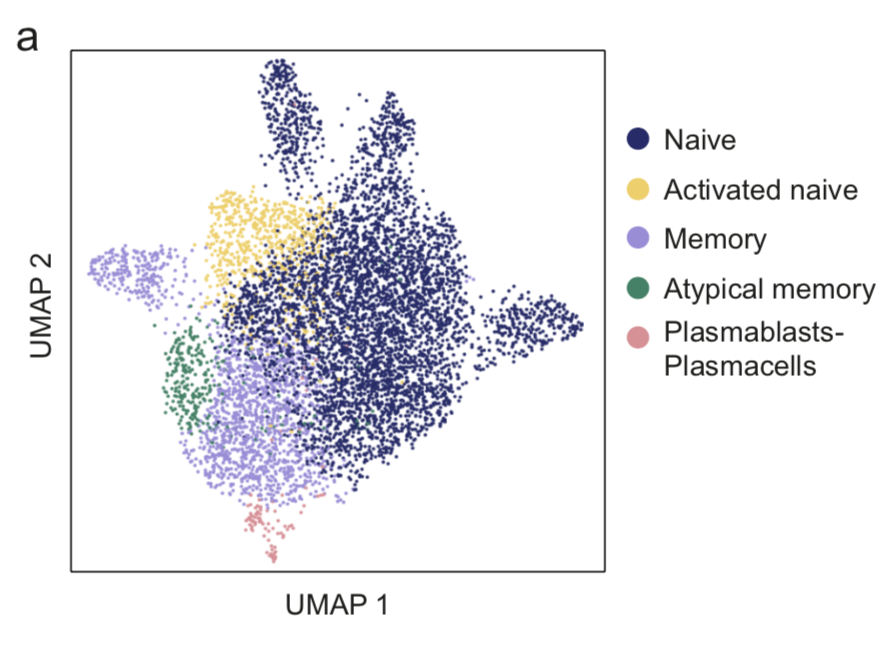
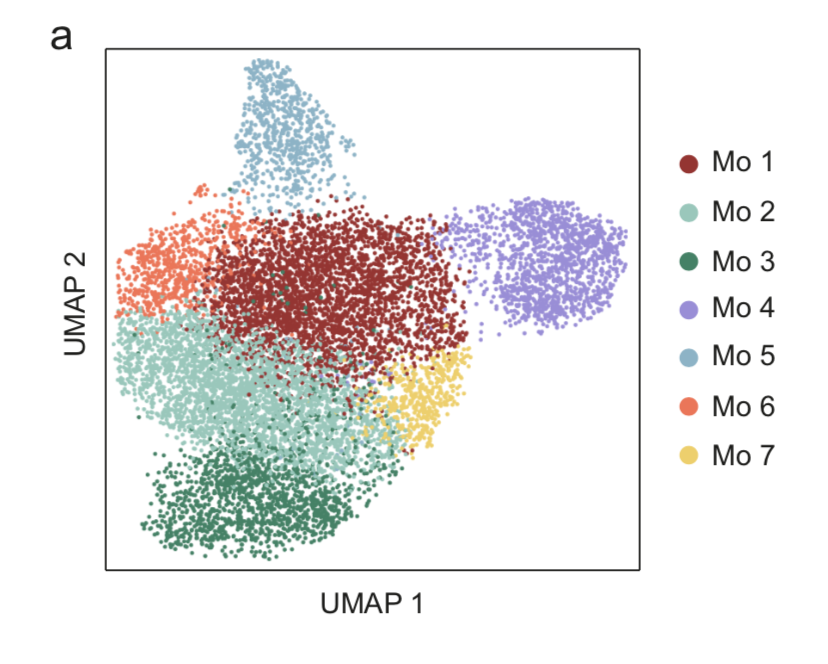
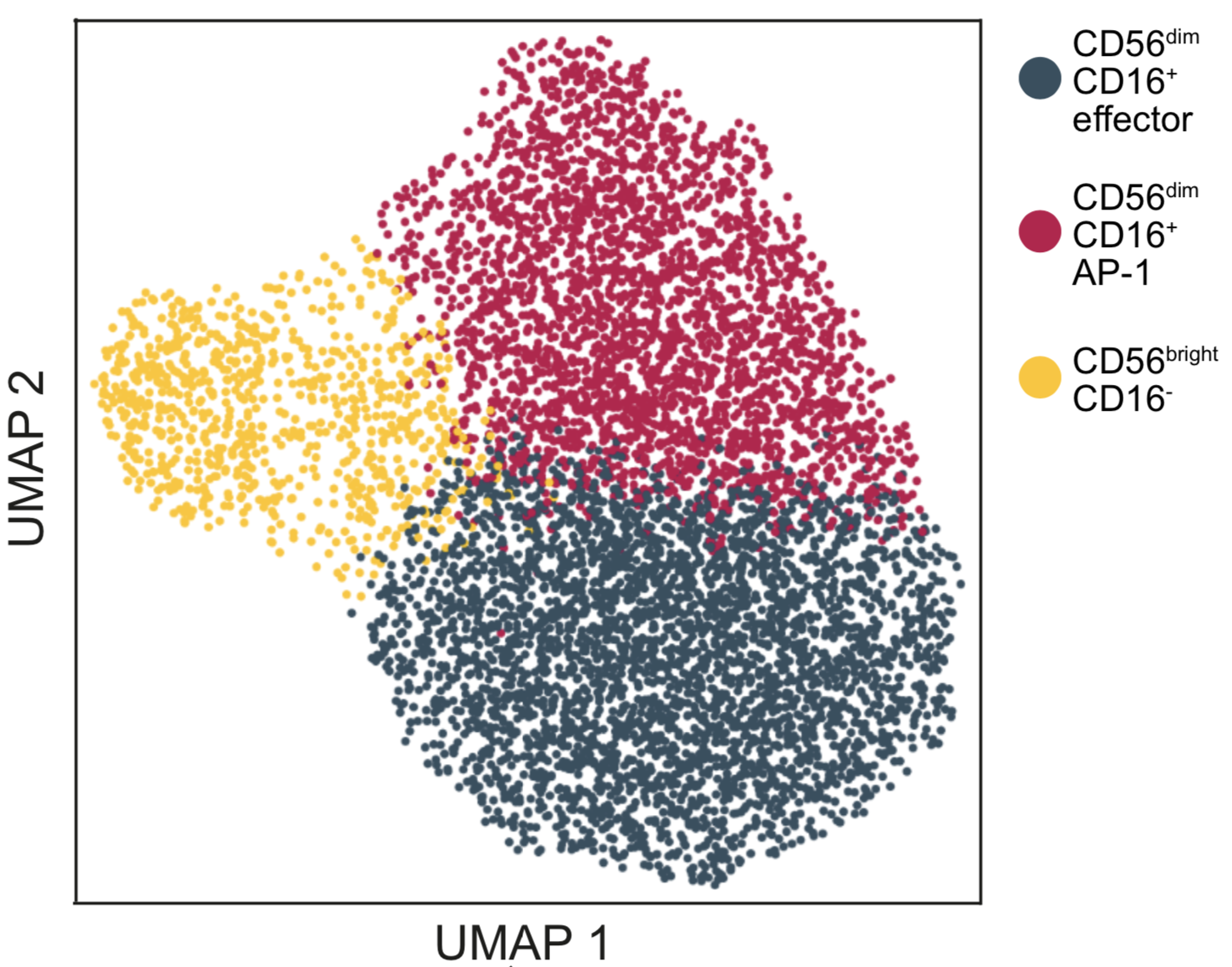
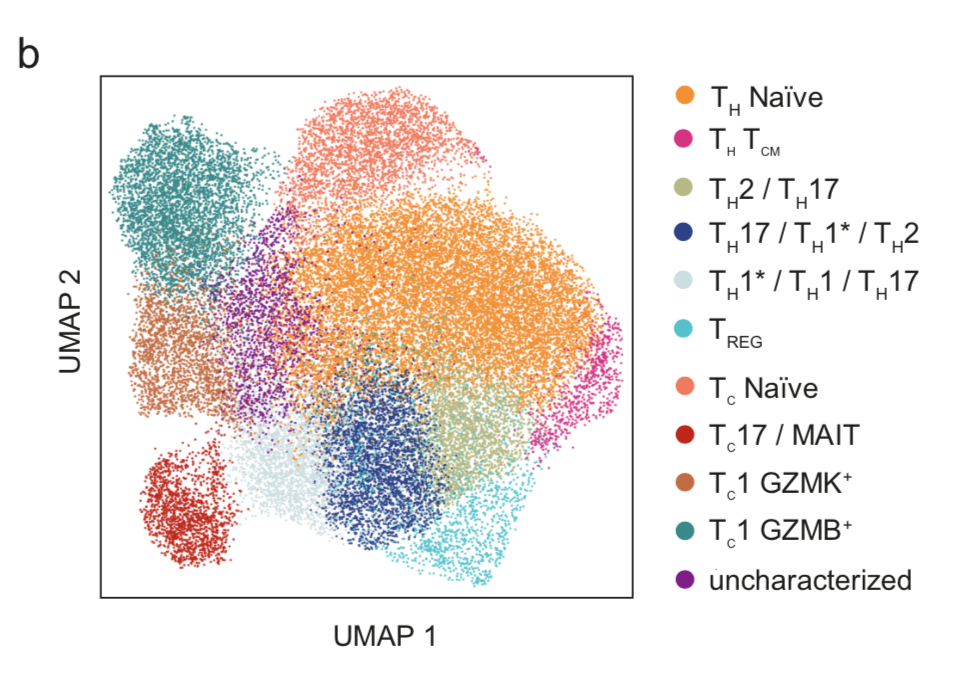
Start exploring the data with iSEE
Click on any of the figures below to start an instance of iSEE with configured for that dataset.
How to explore the data
iSEE delivers a fully interactive browser for the specific single cell dataset it refers to.
Every app starts with a configuration that mirrors the findings presented in the original publication of Notarbartolo et al. (Science Immunology, 2021; Aug 10;6(62):eabg5021) - PMID: 34376481 (https://pubmed.ncbi.nlm.nih.gov/34376481/), DOI: 10.1126/sciimmunol.abg5021 (https://doi.org/10.1126/sciimmunol.abg5021)
In each instance, you can follow a tour that guides you through the main findings as they are presented. At any time, you are free to change any of the defined settings to explore in depth the dataset.